 Baud Lab
Baud Lab
 Systems and Synthetic Biology
Systems and Synthetic Biology
- Group page
- Research lines
- Group members
- Publications

Baud Lab

From October 2021 Junior Group Leader, Centre for Genomic Regulation.
2021-2024 'la Caixa' Junior Leader Fellow, Centre for Genomic Regulation.
2015-2021 Sir Henry Wellcome Postdoctoral Fellow, European Bioinformatics Institute (Cambridge, UK) and University of California San Diego (San Diego, USA).
2013-2015 EMBL Interdisciplinary Postdoctoral Fellow, European Bioinformatics Institute (Cambridge, UK) and EMBL Rome (Rome, Italy).
2009-2013 PhD, Wellcome Trust doctoral training programme in Genomic Medicine and Statistics, University of Oxford (Oxford, UK).
2005-2009 Bachelor’s degree in Biology and Master’s degree in Bioinformatics and Biostatistics, Ecole Normale Superieure (Cachan, France) and Universite Paris XI (Orsay, France).
Summary
How do social partners and the gut microbiome influence health and disease? To address this question, we leverage the fact that many characteristics of social partners and the gut microbiome can be predicted from the genes of social partners and the genes of the gut microbiome, respectively. Thus, instead of trying to measure all the traits of social partners and/or the microbiome that might influence a phenotype of interest, we simply sequence the genes of social partners and/or the genes of the microbiome and model the effect of those genes on the phenotype of interest, measured in focal individuals (Figure 1).
Our laboratory primarily uses laboratory rodents as models but, in the future, we will also investigate indirect genetic effects in humans (e.g. indirect genetic effects from roommates and spouses). Projects in the lab can be purely computational or a combination of experimental and computational work. We have many established collaborations with researchers in Europe and the UK, and in the US.
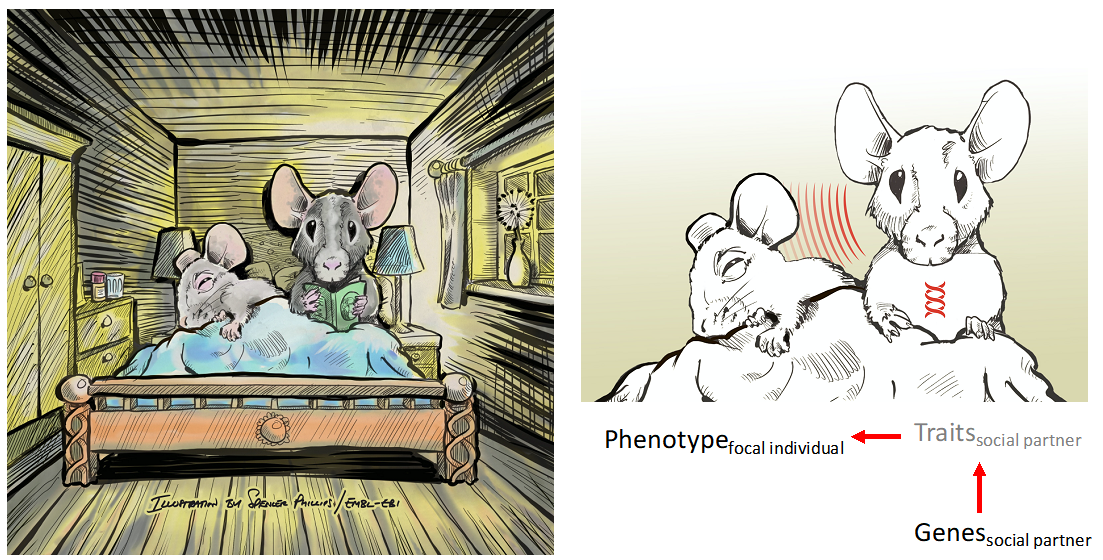
Figure 1. We can study the influence of social partners by sequencing the genes of social partners and modelling their (indirect) effect on the phenotype of interest.
Figure 2. We use genomic technologies and genetic models to dissect the influence of social partners and the gut microbiome on health and disease.
Funding Acknowledgements

The project “Mecanismos de efectos genéticos indirectos que surgen de genotipos de iguals en ratones de laboratorio y en humanos” (PID2021-122651NA-I00) is funded by Agencia Estatal de Investigación (AEI), the Ministerio de Ciencia e Innovación and Fondo Europeo de Desarrollo Regional (FEDER). Project PID2021-122651NA-I00 funded by MCIN/ AEI / 10.13039/501100011033 / FEDER, UE
1. Indirect genetic effects arising from genes of social partners
The phenotype of a focal individual can be affected not only by the individual’s own genotypes (classical “direct genetic effects”) but also by genotypes of social partners (“indirect genetic effects”). Such indirect genetic effects arise when genetic traits of social partners influence the phenotype of interest (Figure 1).

Figure 1.Indirect genetic effects in rodent models.
We have previously shown that many behavioural, physiological and morphological phenotypes measured in laboratory mice are affected by indirect genetic effects arising from genotypes of cage mates (Baud et al., PLOS Genetics 2017) and have developed computational methods to dissect the underlying mechanisms, namely to identify genes and traits of cage mates giving rise to indirect genetic effects (Baud et al., bioRxiv 2020). We will continue to evaluate the impact of indirect genetic effects on biomedical phenotypes and will develop new computational and experimental methods to dissect the underlying mechanisms, in order to better understand how socially-interacting individuals influence each other.
Selected publications:
- A Baud, MK Mulligan, FP Casale, JF Ingels, CJ Bohl, J Callebert, JM Launay, J Krohn, A Legarra, RW Williams, O Stegle (2017) “Genetic variation in the social environment contributes to health and disease”, PLoS Genetics, 13(1): e1006498. DOI:10.1371/journal.pgen.1006498.
The innovative nature and impact of this article was widely recognised (cover of PLOS Genetics, nomination in PLOS Genetics Research Prize 2018 for “scientific excellence and community impact”, citations in high impact journals, interview on BBC Radio 4 Inside Science programme, 34 media reports in 12 countries). - A Baud (2017) “How genetic makeup of a ‘roommate’ can influence health”, PLOS Biologue
https://biologue.plos.org/2017/03/16/understanding-images-how-genetic-ma... - A Baud, FP Casale, J Nicod, O Stegle (2020) “Dissecting the mechanisms underlying indirect genetic effects on biomedical phenotypes: a study of 170 behavioural, physiological and morphological phenotypes measured in adult laboratory mice”, bioRxiv. DOI: 10.1101/302349
2. Indirect genetic effects arising from genes of the gut microbiome
The gut microbiome is emerging as an important factor influencing health and disease, with numerous associations (i.e. correlations) reported between gut microbiome composition and phenotypes of the host. To investigate host-microbiome interactions, we characterise the composition of the gut microbiome by sequencing the 16S ribosomal gene of gut microbes, or their entire genome (metagenomics).
Our lab focuses on microbiome data collected in genetically heterogeneous hosts (laboratory rodents and, in the future, humans). Indeed, we use the genotypes of the host to anchor causal analyses and test whether correlations observed between gut microbiome composition and host phenotype (panel a in Figure 2) reflect causal effects of the microbiome on the host (panel b), causal effects of the host on the microbiome (panel c), or independent effects originating from unknown experimental or environmental factors (panel d).
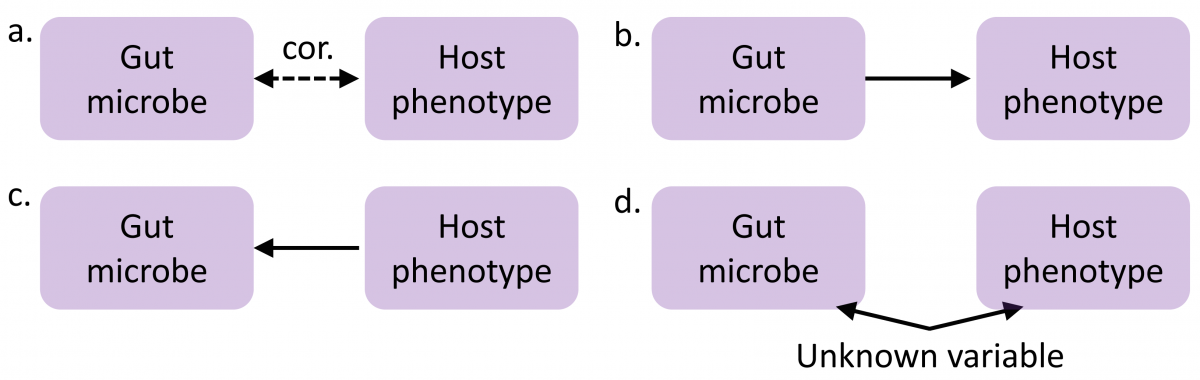
Figure 2. Causal paths underlying host-microbiome associations.
To carry out this work, we are collaborating, in particular, with a US-based consortium of rat geneticists led by Prof. Abraham Palmer (https://ratgenes.org/).

