 Beekman Lab
Beekman Lab
 Genome Biology
Genome Biology
- Group page
- Research lines
- Group members
- Publications

Beekman Lab

July 2023: Group Leader, Centre for Genomic Regulation (CRG, Barcelona, Spain) in double affiliation with the Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
January 2020-June 2023: Group Leader, Centre for Genomic Regulation and Centre for Genomic Analysis (CRG and CNAG-CRG, Barcelona, Spain) in double affiliation with the Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2020-2021: La Caixa Junior Leader Fellow, Centre for Genomic Regulation and Centre for Genomic Analysis (CRG and CNAG-CRG, Barcelona, Spain) in double affiliation with the Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2018-2019: La Caixa Junior Leader Fellow, Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2016-2018: Postdoctoral Fellow, Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2014-2016: Marie Curie Postdoctoral Fellow, Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2013-2014: Rubicon Postdoctoral Fellow, Institute of Biomedical Research August Pi i Sunyer (IDIBAPS, Barcelona, Spain)
2008-2013: PhD in Medicine, Erasmus University (Rotterdam, The Netherlands)
2000-2007: Medical Degree, Erasmus University (Rotterdam, The Netherlands)
2001-2005: MSc in Molecular Medicine, Erasmus University (Rotterdam, The Netherlands)
News
Researchers at the CRG receive €4.5m to study cancer, reproduction and blood formation (10/01/2022)
Renée Beekman will study the impact of translocations, a phenomenon where a chromosome breaks and a portion of it reattaches to a different chromosome, on the formation of tumours. Dr. Beekman’s group will study this in Non-Hodgkin lymphoma to identify potential new vulnerabilities in the disease.
Summary
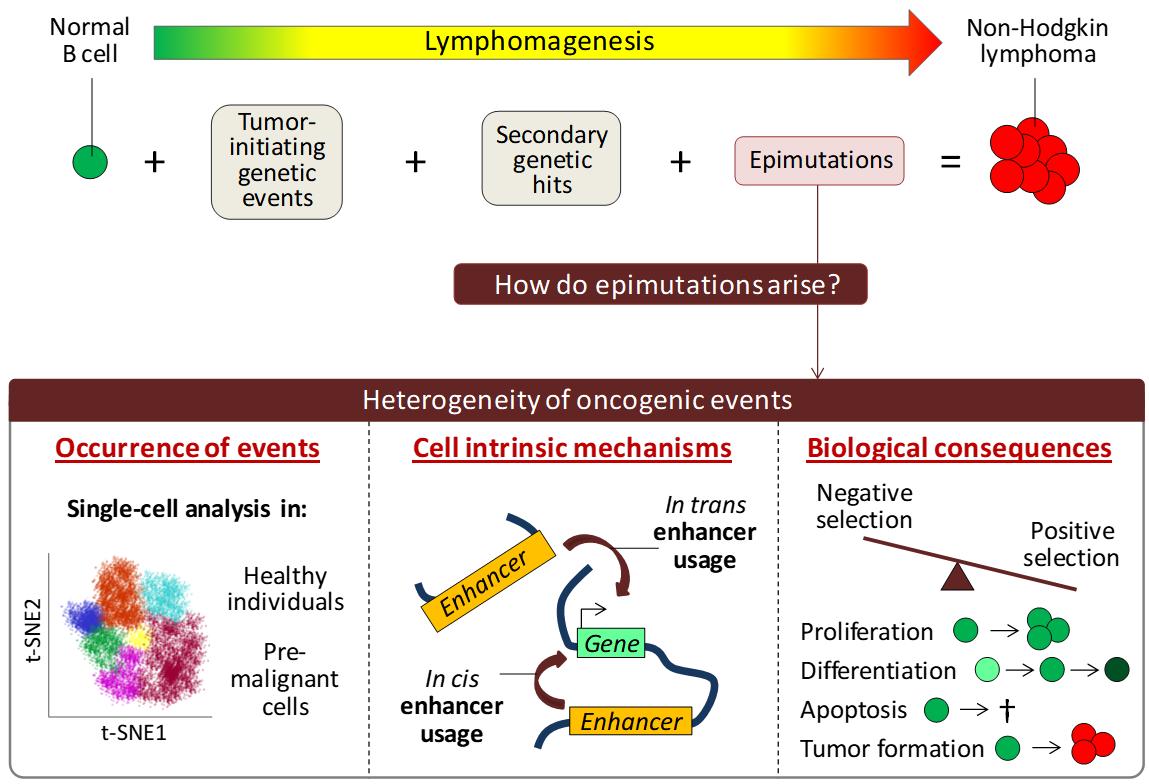
How do malignant cells arise in healthy individuals? Our group aims to shed light upon this question by creating a better understanding of early epigenetic events that contribute to tumorigenesis.
Every tumor originates from a normal cell that, at a given moment, acquires tumor-initiating genetic events, such as translocations and somatic mutations affecting key proto-oncogenes or tumor suppressor genes. These genetic hits turn normal cells into pre-malignant cells, but do not lead to immediate tumor formation. For that, secondary genetic events as well as epigenetic hits, also known as epimutations, are required. In our group we aim to create a better understanding of how epimutations arise and how they contribute to tumorigenesis. We will do this in the context of non-Hodgkin lymphomas, such as mantle cell lymphoma, follicular lymphoma, diffuse large B-cell lymphoma and Burkitt lymphoma.
Fuelled by the advent of single-cell technologies, it is becoming increasingly clear that individual cells in seemingly homogeneous cell populations show cell-to-cell variation in their genetic, epigenetic and gene expression landscapes. We aim to study the heterogeneity of oncogenic events in healthy individuals as well as in pre-malignant cells in vitro (to be created by CRISPR/Cas9 genomic editing) and in vivo using cutting-edge single-cell technologies. On top of that, we aim to define cell intrinsic mechanisms, such as enhancer activation and 3D chromatin interactions, that influence the observed heterogeneity. Altogether, we aim to create new insights into the source of epimutations with the ultimate goal to create a better understanding of the complex process of tumor formation.
Our group is physically located at the Barcelona Biomedical Research Park (PRBB building, C/ Dr. Aiguader, 88, Barcelona, Spain). We are affiliated to the Genome Biology research program (CRG, Barcelona, Spain), and the department of Oncology and Haematology (IDIBAPS, Barcelona, Spain). Together, these provide a unique niche covering basic, translational and clinical biomedical research, and single-cell sequencing technologies. We strongly believe that bringing the knowledge and resources of these different environments together majorly aids to better understand the biology of disease.
Funding Acknowledgements




The project R+D+I project "Plataforma de secuenciación de ADN de lecturas cortas de alto rendimiento y alta flexibilidad" (EQC2021-006925-P) is funded by the Spanish Ministry of Science and Innovation (MICIN), the State Research Agency (AEI) and the "European Union NextGenerationEU/PRTR".
1. Single-cell epigenomics of lymphomagenesis
2. Three-dimensional chromatin structure during early lymphomagenesis
3. Opportunities for interdisciplinary research stays
1. Single-cell epigenomics of lymphomagenesis
In our previous work, we have provided detailed epigenetic characterisations of lymphoid malignancies, with a major focus on mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL) (Queirós A & Beekman R & Vilarrasa-Blasi R et al, Cancer Cell 2016; Beekman R et al, Nature Medicine 2018; Duran-Ferrer M et al, bioRxiv 2020). Importantly, an overarching question in the lymphoma biology field that remains majorly unexplored so far is: “How do normal lymphocytes transform into lymphoma cells and (besides genetic lesions) to what extent do epigenetic events drive this process?” We have started projects focusing on this area of study to address the following specific questions: “In which order do genetic and epigenetic events accumulate during early lymphoma formation?” and “Does epigenetic mosaicism exist in healthy individuals and does it play a role in lymphomagenesis?” We shall answer these questions, using MCL as disease model, by performing single-cell analysis of normal cells that suffer tumor onset and identifying and characterising pre-malignant stages that arise during early lymphoma formation.
- Beekman R, Chapaprieta V, Russiñol N, Vilarrasa-Blasi R, Verdaguer-Dot N, Martens JHA, Duran-Ferrer M, Kulis M, Serra F, Javierre BM, Wingett SW, Clot G, Queirós AC, Castellano G, Blanc J, Gut M, Merkel A, Heath S, Vlasova A, Ullrich S, Palumbo E, Enjuanes A, Martín-García D, Beà S, Pinyol M, Aymerich M, Royo R, Puiggros M, Torrents D, Datta A, Lowy E, Kostadima M, Roller M, Clarke L, Flicek P, Agirre X, Prosper F, Baumann T, Delgado J, López-Guillermo A, Fraser P, Yaspo ML, Guigó R, Siebert R, Martí-Renom MA, Puente XS, López-Otín C, Gut I, Stunnenberg HG, Campo E, Martin-Subero JI. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018 Jun;24(6):868-880.
- Duran-Ferrer M, Clot G, Nadeu F, Beekman R, Baumann T, Nordlund J, Marincevic-Zuniga Y, Lönnerholm G, Rivas-Delgado A, Martin S, Ordoñez R, Castellano G, Kulis M, Queirós A, Seung-Tae L, Wiemels J, Royo R, Puiggrós M, Lu J, Gine E, Beà S, Jares P, Agirre X, Prosper F, López-Otín C, S. Puente X, Oakes CC, Zenz T, Delgado J, López-Guillermo A, Campo E, Martin-Subero JI. The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome. bioRxiv 2020.02.06.937383.
- *Queirós A, Beekman R*, Vilarrasa-Blasi R*, Duran-Ferrer M, Clot G, Merkel A, Raineri E, Russiñol N, Castellano G, Beà S, Navarro A, Kulis M, Verdaguer-Dot N, Jares P, Enjuanes A, Calasanz MJ, Bergmann A, Vater I, Salaverría I, van de Werken HJG, Wilson WH, Datta A, Flicek P, Royo R, Martens J, Giné E, Lopez-Guillermo A, Stunnenberg HG, Klapper W, Pott C, Heath S, Gut IG, Siebert R, Campo E, Martín-Subero JI. Decoding the DNA methylome of mantle cell lymphoma in the light of the entire B cell lineage. Cancer Cell. 2016 Nov 14;30(5):806-821. *Shared first authors.
2. Three-dimensional chromatin structure during early lymphomagenesis
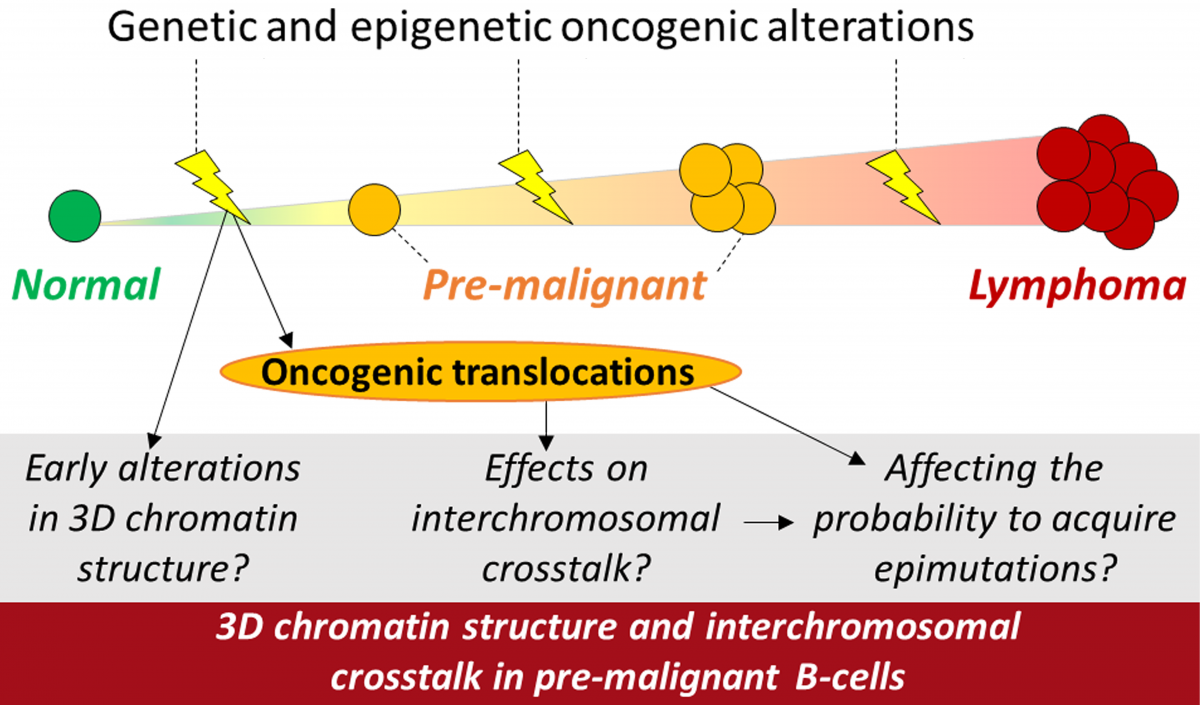 In our previous work we have performed detailed analyses of the three-dimensional (3D) chromatin structure in normal and malignant B cells both at the genome-wide level and at specific (proto-onco)genic loci (Puente XS et al, Nature 2015; Queirós A & Beekman R & Vilarrasa-Blasi R et al, Cancer Cell 2016; Beekman R et al, Curr Opin Hematol 2018; Vilarrasa-Blasi R & Soler-Vila P et al, bioRxiv 2019). It remains to be elucidated, however, how the 3D chromatin structure gets altered during tumor onset and how these alterations play a role in early lymphomagenesis. We have started projects focusing on this research area aiming to answer the following specific questions: “Do oncogenic translocations affect the epigenetic heterogeneity of pre-malignant cell pools by altering interchromosomal crosstalk?” and, “Does this process result in increased occurrence of epimutations that drive tumor formation?”. We shall address these questions by studying different facets of interchromosomal crosstalk in pre-malignant lymphocytes carrying genetically engineered lymphoma-associated genetic translocations.
In our previous work we have performed detailed analyses of the three-dimensional (3D) chromatin structure in normal and malignant B cells both at the genome-wide level and at specific (proto-onco)genic loci (Puente XS et al, Nature 2015; Queirós A & Beekman R & Vilarrasa-Blasi R et al, Cancer Cell 2016; Beekman R et al, Curr Opin Hematol 2018; Vilarrasa-Blasi R & Soler-Vila P et al, bioRxiv 2019). It remains to be elucidated, however, how the 3D chromatin structure gets altered during tumor onset and how these alterations play a role in early lymphomagenesis. We have started projects focusing on this research area aiming to answer the following specific questions: “Do oncogenic translocations affect the epigenetic heterogeneity of pre-malignant cell pools by altering interchromosomal crosstalk?” and, “Does this process result in increased occurrence of epimutations that drive tumor formation?”. We shall address these questions by studying different facets of interchromosomal crosstalk in pre-malignant lymphocytes carrying genetically engineered lymphoma-associated genetic translocations.
- Beekman R, Amador V, Campo E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr Opin Hematol. 2018 Jul;25(4):299-306.
- Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, Munar M, Rubio-Pérez C, Jares P, Aymerich M, Baumann T, Beekman R, Carrio A, Castellano G, Clot G, Colado E, Colomer D, Costa D, Delgado J, Enjuanes A, Estivill X, Ferrando AA, Gelpí JL, González B, González S, González M, Guigó R, Gut M, Hernández-Rivas JM, López-Guerra M, Martín-García D, Navarro A, Nicolás P, Orozco M, Payer AR, Pinyol M, Pisano DG, Puente DA, Queirós AC, Quesada V, Romeo-Casabona CM, Royo C, Royo R, Rozman M, Salaverría I, Stamatopoulos K, Tamborero D, Terol MJ, Valencia A, López-Bigas N, Torrents D, Gut I, López-Guillermo A, López-Otín C, Campo E. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015 Oct 22;526(7574):519-24.
- *Queirós A, Beekman R*, Vilarrasa-Blasi R*, Duran-Ferrer M, Clot G, Merkel A, Raineri E, Russiñol N, Castellano G, Beà S, Navarro A, Kulis M, Verdaguer-Dot N, Jares P, Enjuanes A, Calasanz MJ, Bergmann A, Vater I, Salaverría I, van de Werken HJG, Wilson WH, Datta A, Flicek P, Royo R, Martens J, Giné E, Lopez-Guillermo A, Stunnenberg HG, Klapper W, Pott C, Heath S, Gut IG, Siebert R, Campo E, Martín-Subero JI. Decoding the DNA methylome of mantle cell lymphoma in the light of the entire B cell lineage. Cancer Cell. 2016 Nov 14;30(5):806-821. *Shared first authors.
- Vilarrasa-Blasi R, Soler-Vila P, Verdaguer-Dot N, Russiñol N, Di Stefano M, Chapaprieta V, Clot G, Farabella I, Cuscó P, Agirre X, Prosper F, Beekman R, Beà S, Colomer D, Stunnenberg HG, Gut I, Campo E, Marti-Renom MA, Martin-Subero JI. Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. bioRxiv 764910.
3. Opportunities for interdisciplinary research stays
Are you trained in, or are you receiving training in (i) medicine, (ii) mathematics, (iii) informatics or related disciplines and would you like to be exposed to the field of biomedical research? Please let us know! Besides providing opportunities to highly motivated students in the fields of biology, biomedicine and bioinformatics, our group is particularly interested in exploring possibilities to offer short stays to individuals with expertise in the complementary disciplines listed above. We believe that the mutual exchange of knowledge during these stays is key to build bridges between research fields, providing the soil for fruitful interdisciplinary collaborations and consequent scientific breakthroughs.



